Project 1: Xenon-129 NMR Biosensing
Magnetic resonance imaging (MRI) is one of the most used and versatile medical imaging techniques. In many respects, MR spectroscopy and imaging are ideal for in vivo molecular imaging, due to the ability to generate non-invasive, high-resolution images of human anatomy. The sensitivity of atomic nuclei to their local environment provides useful molecular information. The radio frequencies required to interrogate samples by MRI are non-damaging and completely compatible with living specimens. However, despite the abundant proton signals (from water, fat, etc.), 1H MRI typically cannot reach desired detection sensitivity for early-stage cancer diagnostics. MRI contrast agents that improve image contrast are typically limited to micromolar-to-millimolar detection sensitivity.
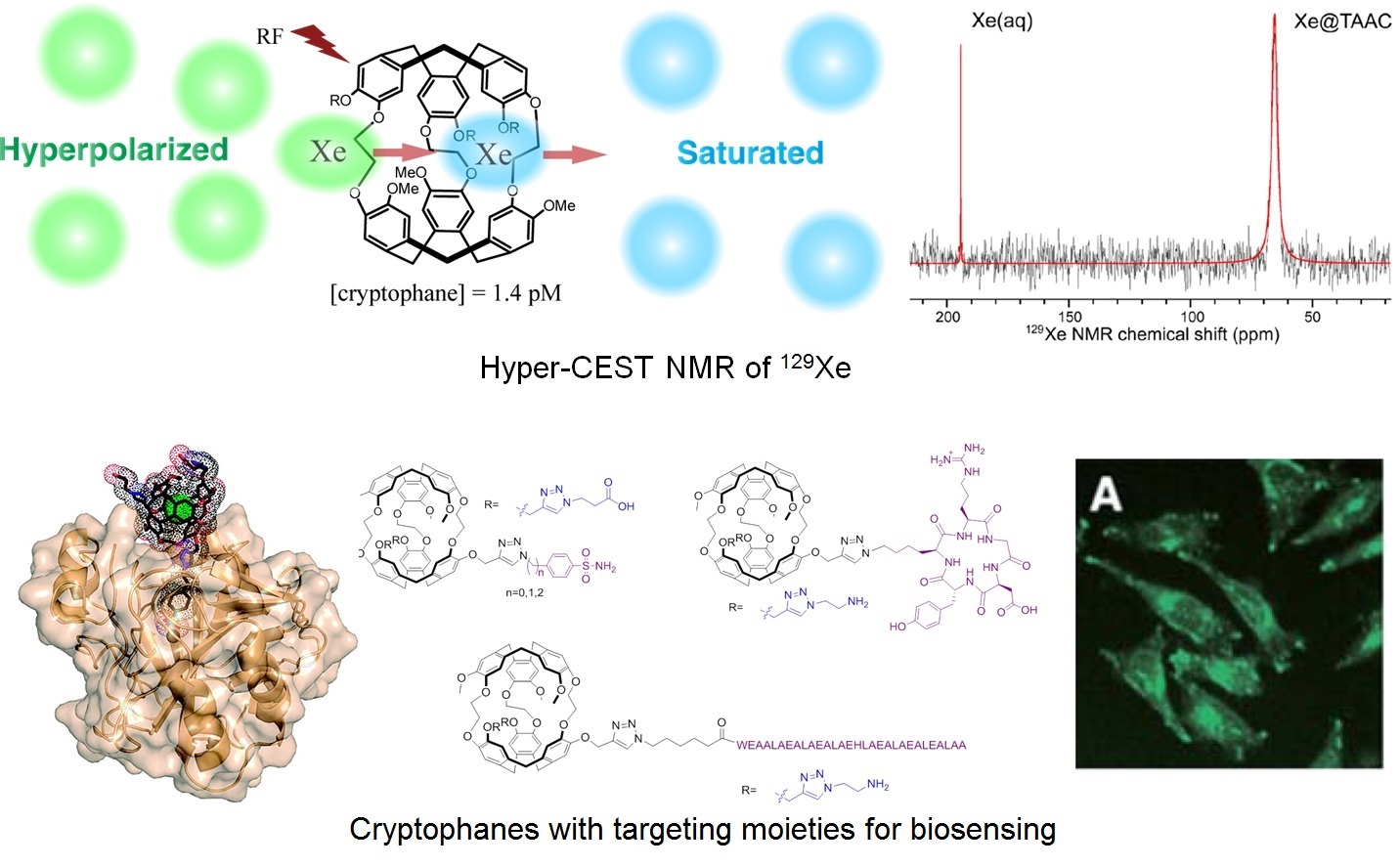
This sensitivity barrier can be overcome through the use of hyperpolarized MRI. This form of imaging exploits nuclei that can be hyperpolarized – prepared in nonequilibrium states of artificially high spin polarization – and introduced into the body by injection or inhalation. One particularly useful nucleus is 129Xe, as it has ˝ nuclear spin, a wide chemical shift window, and inert chemical properties. The significant polarizability of xenon contributes to its significant water solubility, as well as high affinity for void spaces in small molecules and materials. Moreover, greater than 100,000-fold NMR/MRI signal enhancement is achievable by state-of-art hyperpolarization (nuclear spin polarization) instruments. Combining a targeting moiety with a Xe host molecule serves the purpose of delivering Xe to a specific site, as well as generating a specific 129Xe NMR chemical shift. The goal of these studies is to provide molecular signatures that distinguish cancer cells from healthy cells. Our focus is on the early and accurate diagnosis of cancers of the lungs and brain, where high concentrations of hyperpolarized 129Xe can be readily delivered via inhalation.
Thus, our lab is developing various 129Xe-cryptophane biosensors targeting cancer biomarkers such as carbonic anhydrase (1) and integrin receptor (2). In addition, we are developing pH-sensitive and other “smart” cryptophane hosts, which could expand the application of Xe-cryptophane biosensing (3). In collaboration with the Christianson lab, we are working to assign specific 129Xe NMR chemical shifts to particular carbonic anhydrase-cryptophane interactions identified by high-resolution X-ray structure determinations.
A final area of research exploits the rapid chemical exchange of Xe between “bound” (i.e., cryptophane) and “free” (i.e., bulk water) states to achieve even greater detection sensitivity. This has led to our lab’s implementation and refinement of the hyperpolarized 129Xe chemical exchange saturation transfer (Hyper-CEST) NMR technique first reported by the Pines lab, which produces an additional 105-fold sensitivity enhancement with our in-house instrument. In tandem with cryptophane studies, we are investigating different biological structures for their ability to produce a useful Hyper-CEST effect.
Project 2: Light-Activated Oligonucleotides
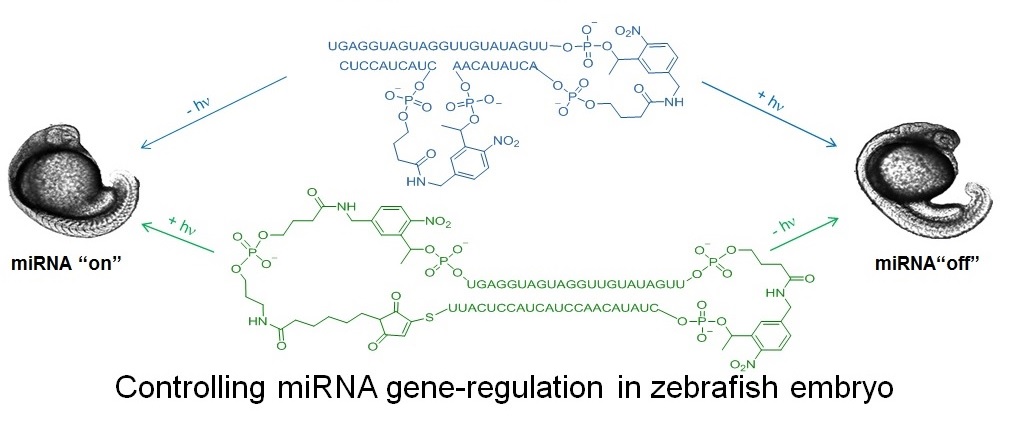
In order to study gene expression with greater spatial and temporal resolution, our lab has developed photochemical tools for controlling mRNA translation, allowing protein expression to be both turned “on” or “off” with light. We typically design light-activated (“caged”) oligonucleotides whose binding to target mRNA changes upon near-UV or visible irradiation. We have recently developed methods for controlling microRNA (miRNA) with light in zebrafish embryos. Because a single miRNA can regulate multiple gene targets, it is important to develop tools to provide a way of elucidating the function of a miRNA in different locations and times. Two caged constructs have been developed allowing let-7, a zebrafish miRNA of broad biological importance, to be switched from both "off to on" and "on to off" with 365 nm light (1). Efforts are underway to make caged oligos whose activity changes with near-IR excitation.
Also under development are photochemical methods for isolating mRNA from single cells in buried brain tissues. Our initial work in this area has focused on analyzing the diverse transcriptomes of neurons. In collaboration with the Eberwine lab, we have developed Transcriptome In Vivo Analysis (TIVA)-tags, which incorporate photocleavable spacers linking blocking strands to a poly-U probe targeting the poly-A tail of mRNA. This design allows as much of the probe as possible to be blocked while maintaining low melting temperatures post-photolysis. TIVA-tags are functionalized with a Cy3/Cy5 FRET pair to monitor dissociation of the oligonucleotide in vivo and in vitro, and a cell penetrating peptide to assist with cell uptake. Finally, a biotin moiety attached to the oligonucleotide is utilized to pull down the mRNA with a streptavidin bead after the TIVA-tag is uncaged. TIVA-tags have been used successfully to isolate mRNA from single neurons both in culture and in live mouse hippocampal slices.
Project 3: Synergistic Protein-Nanoparticle Interactions
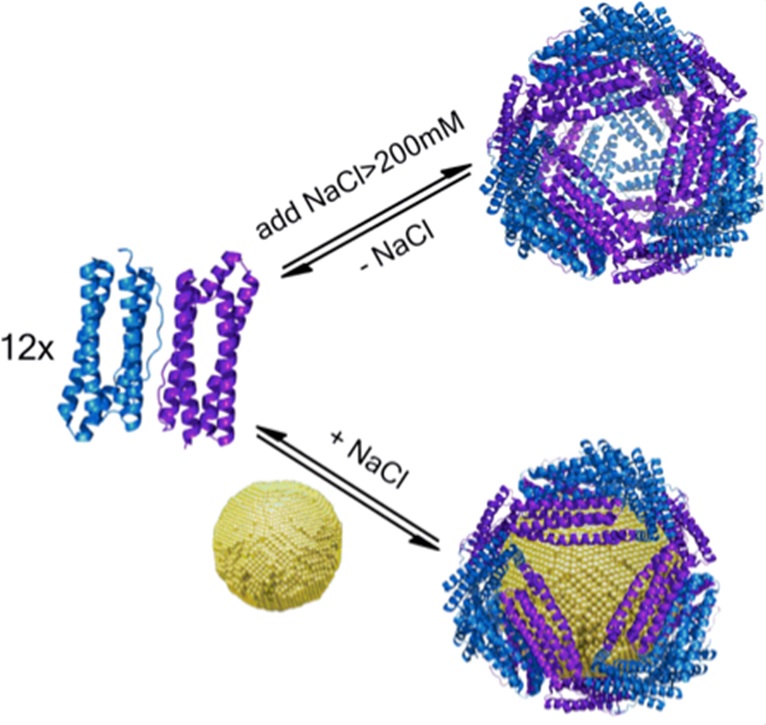
Nanoparticles have many unique optical, electronic, and magnetic properties that would be highly useful in many biological applications. However, current understanding of how nanoparticles interact with biomolecules, particularly proteins, is limited. This project seeks to expand that understanding using ferritin from the archaeon A. fulgidus as a model system. This ferritin is unique in that it can assemble and disassemble from its 24-mer cage structure into dimers, depending on the salt concentration. We have evidence that the protein can also self-assemble in the presence of 5-10-nm diameter gold nanoparticles, even under low salt conditions where the protein is normally in dimer form. We have shown this by circular dichroism spectroscopy, transmission electron microscopy, native gel electrophoresis, size exclusion chromatography, and fluorescence spectroscopy. We are currently using cryo-electron microscopy and fluorescence correlation spectroscopy (in collaboration with the Gai lab) to investigate further the ferritin-gold nanoparticle structure and assembly process, respectively. We are also looking at the protein assembly around nanoparticles of different compositions and shape in collaboration with the Murray lab. We employ the tools of protein design and protein crystallography, as well as a wide range of nanoparticle characterization methods to study these bio-nano assemblies. Our work is leading to the development of a protein toolkit for achieving nanoparticle stabilization, nano-separations, templating of inorganic reactions, as well as control of catalytic reactions at metal surfaces mediated by the protein shell.
Project 4: Fluorescent Anesthetic Probes
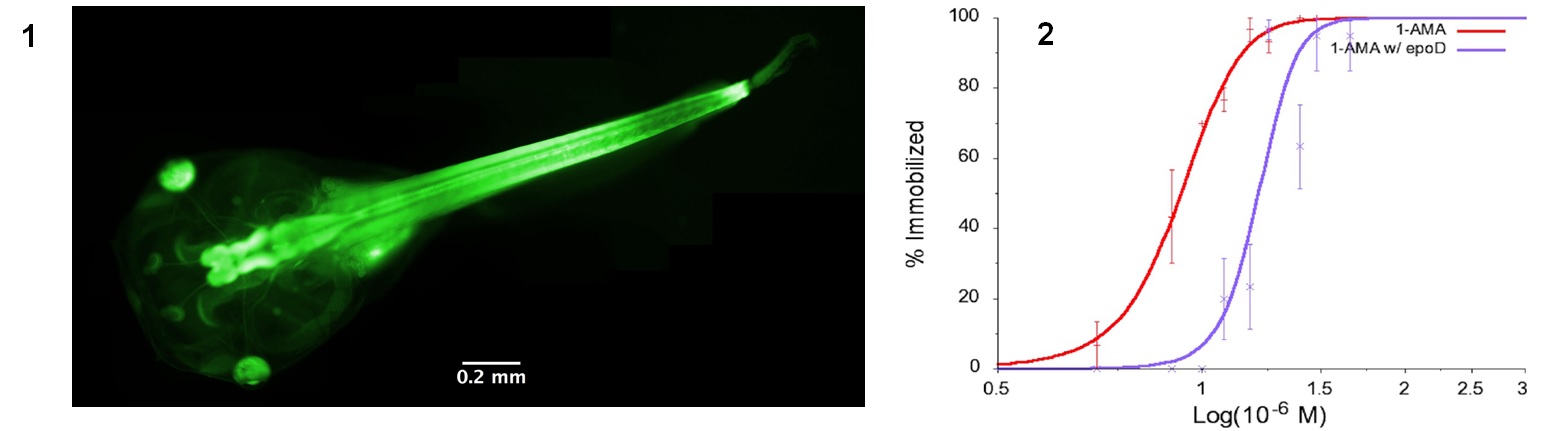
While general anesthetics have been used in medicine for more than 170 years, it is still unknown how these drugs function at the molecular level, which hinders the development of more potent and safer anesthetics. In collaboration with Rod Eckenhoff from the UPenn School of Medicine Department of Anesthesiology and Critical Care, we have identified the first fluorescent general anesthetic, 1-aminoanthracene (1-AMA). It was shown to immobilize reversibly zebrafish and tadpoles, potentiate GABAergic transmission, and localize to neuronal tissue (1). To further identify the molecular targets and binding sites, we made the photoactive analog 1-azidoanthracene (1-AZA), which generates a reactive nitrene upon UV photolysis and crosslinks with the protein targets. Combining SDS-PAGE and tandem mass spectrometry, it was identified that the most abundant crosslinked protein by 1-AZA is beta-tubulin. Using two microtubule stabilizing agents epothilone-D and discodermolide (2), we further substantiated that the neuronal microtubules are an “on-path” target of anthracene general anesthetics. Additional studies are underway to develop new fluorescent anesthetic probe molecules, and identify other important protein targets.
Project 5: Macromolecules at the Air-Water Interface
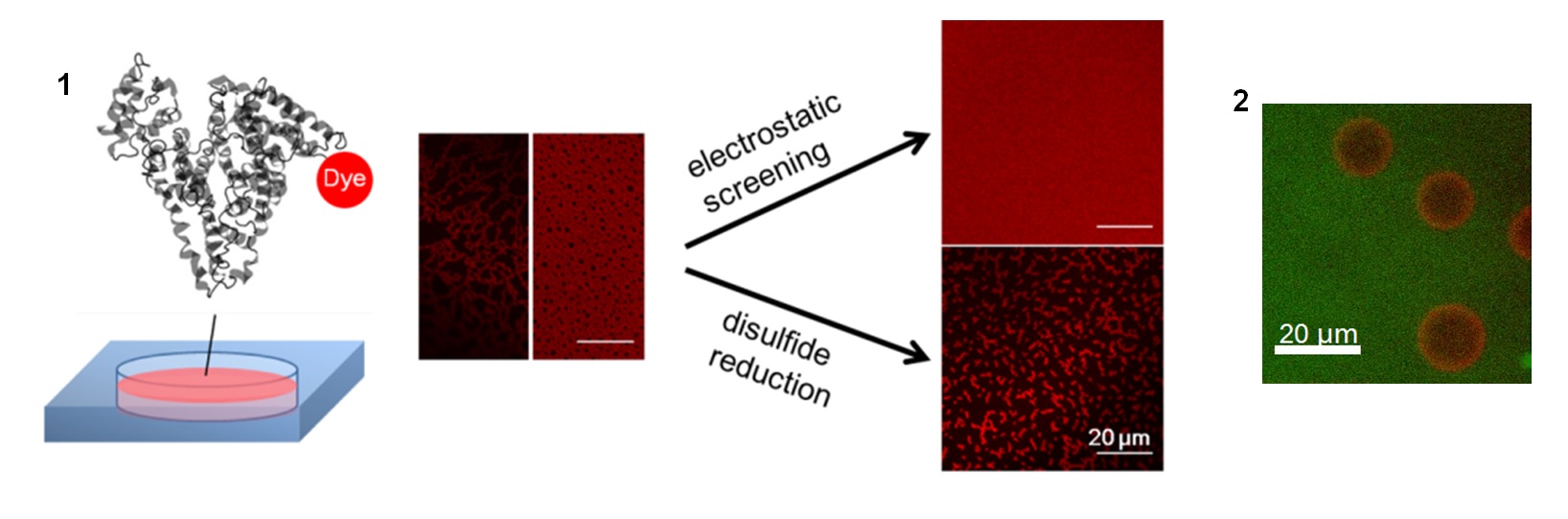
Controlling the adsorption and configuration of proteins at the air-water interface (AWI) is important to many biotechnological and biomedical processes, including gas embolism. nBy chemically labeling proteins with minimally perturbing chromophores, we utilize fluorescence imaging to obtain qualitative and quantitative information of proteins at the AWI, such as interfacial protein concentration, adsorption kinetics, nano- to micro- scale morphology, and interaction with other polymeric molecules. Our previous study showed that solution conditions such as ionic strength and reducing agents can alter the assembly structure of bovine serum albumin at the AWI (1). Recently, we studied the interaction at the AWI between a common plasma protein immunoglobulin G and polydimethysiloxane (PDMS). Confocal laser scanning microscopy revealed distinct microscale structures formed in this protein-polymer system (2).